A Python Demo for MD simulation of Lennard-Jones Fluid in NVT Ensemble
一个简单的demo,用Python实现在NVT系综中Lennard-Jones流体的分子动力学模拟。原理见Molecular Dynamics in NVT ensemble一文。
Code: 参数设置和变量初始化
1 | |
Code: 单步积分器
1 | |
力与势能的计算
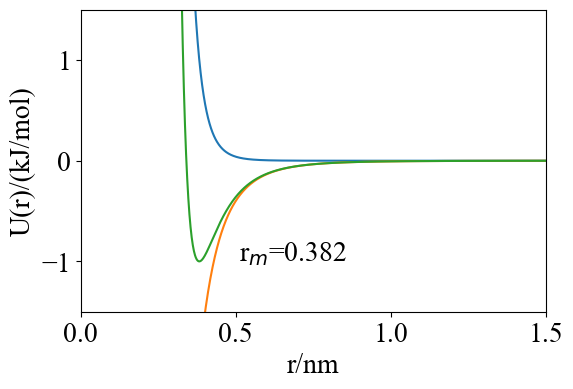
- Lennard-Jones Potential
\[ U^{LJ}(r_{ij})=4\epsilon \bigl(\frac{\sigma^{12}}{r_{ij}^{12}}-\frac{\sigma^6}{r_{ij}^6}\bigr)\\ r_{ij}=\sqrt{(x_i-x_j)^2+(y_i-y_j)^2+(z_i-z_j)^2} \]
Force \[ Fx_i=-\frac{\partial U^{LJ}(r_{ij})}{\partial x_i}=4\epsilon \bigl(\frac{12\sigma^{12}}{r_{ij}^{13}}-\frac{6\sigma^6}{r_{ij}^7}\bigr)\cdot\frac{x_i}{r_{ij}} \] Extreme point \[ r_m=\sqrt[6]{2}\,\sigma \]
1 | |
1 | |
参数设置
Simulation parameters: the time step (ps): 0.01 the proportion that dt/dt(NHC): 20 the number of the simulation steps: 10000 the number of particles: 216 the target temperature (K): 119.8 the number of thermostats: 2 the character time scale \(\tau\) (ps): 0.2
Boundary Condition: cubic box lbox= 2.04 nm
Topology: 216 Ar atoms, mass 39.94 g/mol LJ_epsilon: 0.99607 kJ/mol LJ_sigma: 0.3405 nm Cutoff radial: 1.0 nm
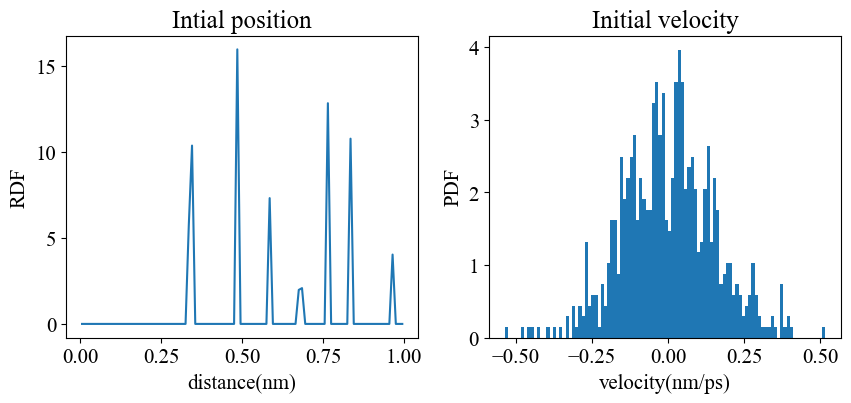
Initialization: positions: put the particles on the cubic lattice velocities: use Box-Miller method sampling the MB distribution at Ti=110 K thermostat variables: zeros
Dump the T,UE,KE,velocities and positions every step!
Code:模拟与数据存储
1 | |
Set up the simulation paramaters
the type of periodic boundary condition (e.g. 'cubic','rectangle','noPBC'): cubic
the length of the cubic box (nm): 2.04
Simulation start!
[0, 110, 296.327429244, -789.1019846977582]
... ...
Simulation finish!1 | |
1 | |
数据分析与可视化
径向分布函数 (RDF)
- 原子b对原子a的径向分布函数定义为:
\[ g_{ab}(r)=\frac{P_b(r)}{4\pi\rho_br^2dr} \]
其中\(P_b(r)\)是原子b出现在离原子a距离为\([r,r+dr)\)的球形壳层中的概率。故径向分布函数就是径向概率密度除以原子b的平均密度,归一化到\(N_b\)。在无限远处 \(g_{ab}(\infty)=\rho_b\)。 实际计算时,首先设定最大距离 \(r_{max} (<\frac{1}{2}L_{box})\) 和区间数 \(N_r\) 。将 \([0,r_{max})\) 分为宽度为\(\Delta r=r_{max}/N_r\) 的 \(N_r\) 个小区间,对轨迹的每一帧、每个a、b原子对,统计距离落在区间 \([r_i,r_i+dr)\) 的频数 \(h_{ab}(i)\)。可用下式获取小区间的index: \[ i=int(\frac{r}{\Delta r}) \] 最终径向分布函数可用下式计算: \[ g_{ab}(r_i)=\frac{h_{ab}(r_i)}{4\pi\rho_br_i^2\Delta rN_{conf}N_a} \]
1 | |
初始条件
1 | |
模拟结果
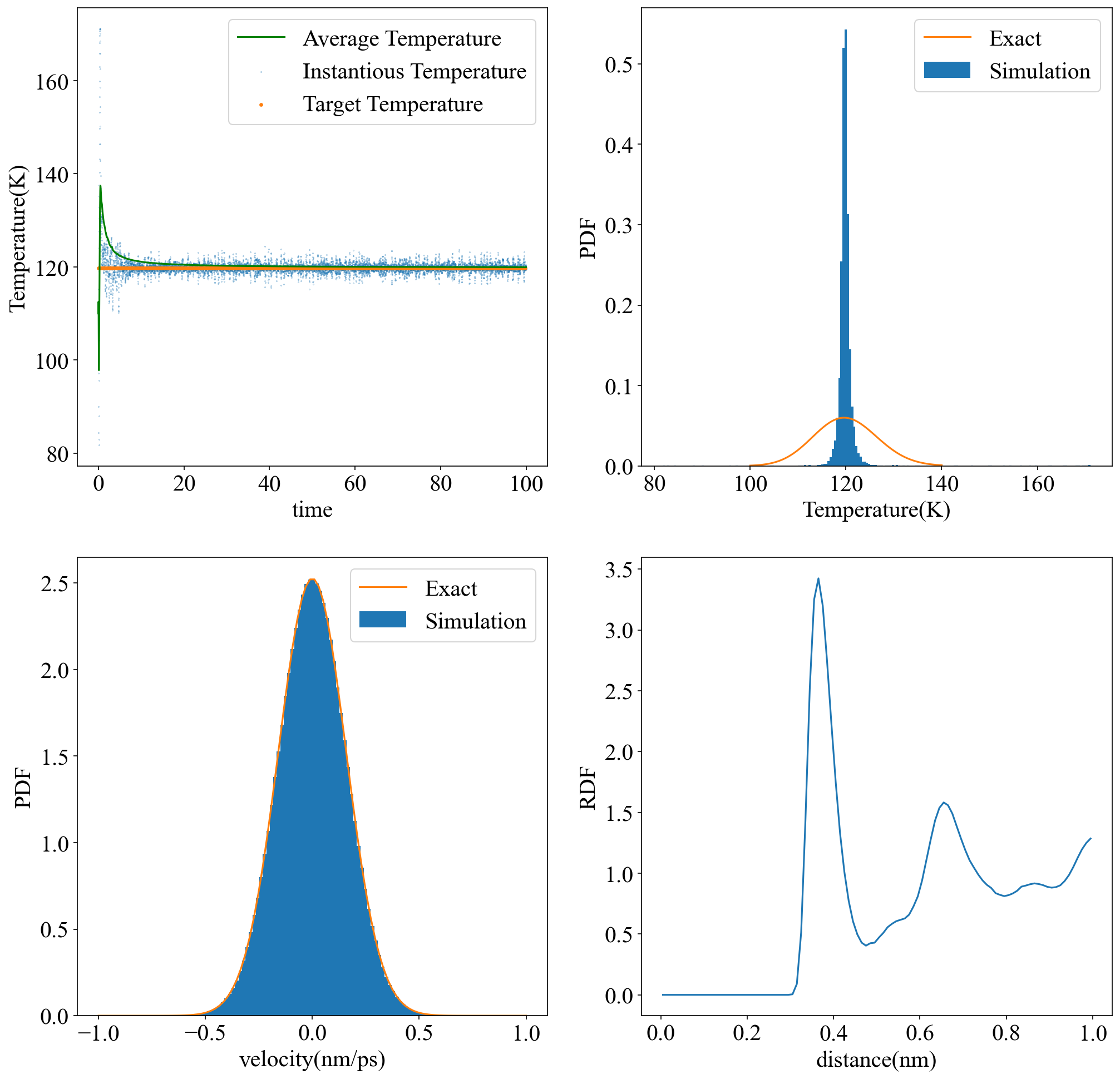
- 20 ps 后平均温度稳定在目标温度附近,速度分布与MB分布吻合得很好
- 经典离域子体系理想的温度分布为:
\[ f(T)=\frac{1}{\Gamma(\frac{3N+1}{2})}(\frac{3N}{2T_0})^{\frac{3N+1}{2}}\cdot T^{\frac{3N-1}{2}}\cdot exp(-\frac{3N}{2T_0}T) \]
当N很大时,上述分布区域Gauss分布,且变得尖锐。温度的相对涨落满足: \[ \frac{\sigma^2_T}{<T>^2}=\frac{2}{3N} \] 由图可见,虽然任一自由度的速度分布与理想分布十分吻合,模拟产生的温度分布相比理想情形过于尖锐。调节热浴质量参数可能使温度涨落更接近理想。
- 径向分布函数在LJ势的极小值点附近出现第一个极大值,与实际情况吻合。由于模拟时长较短,曲线不够平滑。最大距离不能超过模拟盒子的一半长度,以免计算粒子与自身镜像的作用,因而此处无法显示 1.5 nm 附近RDF趋于1的情况。
1 | |
参考文献
Mark E. Tuckerman. (2010). Statistical Mechanics: Theory and Molecular Simulation. Oxford: Oxford University Press.
Daan Frenkel & Berend Smit著, 汪文川译. (2002). 分子模拟:从算法到应用. 化学工业出版社.